Preface
Biological information flows in the direction of "DNA → mRNA → protein → cells → organ → individual → group". The interaction of gene and environment is the root cause of all things in the world. If the human genome program opens the door to thinking where we come from, sequencing technology has subverted our traditional understanding of the world, so that researchers may discuss the problems that could not be explored in the past, and provide a useful perspective for human beings to be closer to the essence of nature and their mysteries.
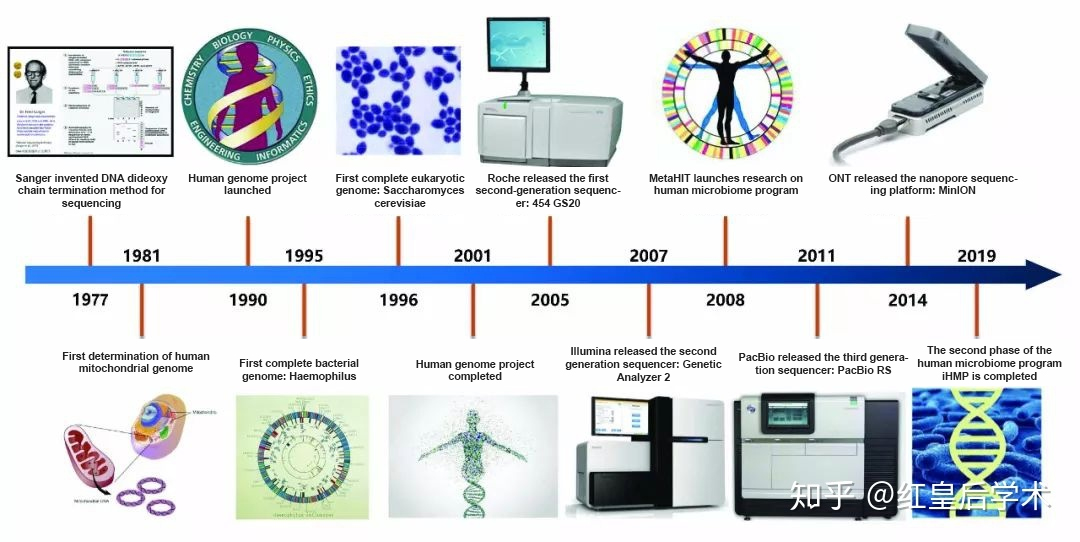
Figure 1 Development of Gene Sequencing Technology, Source: Red Queen Academic
The application of high-throughput sequencing technology in the field of human medicine research mainly includes: whole genome sequencing, total exon group sequencing, target area sequencing and other technologies. The high-throughput sequencing technology is mainly through DNA fragments, library construction, cross linked amplification of library and vector, and sidewalk sequencing reactions on the carrier surface to obtain genetic information on individual or group. Future research will adopt larger sample groups, combined with statistically reliable related analysis, and gradually build a complete genetic mutation database to help scientific researchers better understand the nature of life, provide doctors with guidance on prevention, diagnosis and treatment.
Whole Genome Sequence
1. Technical Principle
Whole Genome Sequencing (WGS) refers to the technical methods of using high-throughput sequencing platform to sequence the whole genome of different human individuals or populations, and to analyze biological information at the individual or population level. Whole genome sequencing can comprehensively excavate the genetic variation of DNA levels, including large structural variations, providing important information for screening diseases and susceptible genes, studying onset and genetic mechanisms, and inferring population migration and evolution.
The whole genome sequencing can be mainly divided into:
De novo sequencing: The genome of a species can be sequenced without any reference genome information, and the genome sequence map of the species can be obtained by splicing and assembling with bioinformatics analysis methods, so as to promote the follow-up research of the species. Only after de novo sequencing can a detailed genetic analysis be carried out on any organism, which is often used for genetic analysis of highly mutated viruses and other frequently mutated species for subsequent research on this species. Only after de novo sequencing can a detailed genetic analysis be carried out on any organism, which is often used for genetic analysis of highly mutated viruses and other frequently mutated species.
Re-sequencing: It is a genome sequence of different individuals with reference genome species, and the analysis of differences between individuals or populations on this basis. After the existing genome is published, re-sequencing can be directly performed on sequenced reads without splicing. In short, the genome sequence is performed on the self-known species of genomic sequences. Its essence is to find variations-mainly used to assist researchers to find variations such as single nucleotide polymorphism (SNPs), copy number variation (CNV) and indel.
Compared with other sequencing forms of human genome, the platform used for whole genome sequencing needs to have a large throughput. For example, the HiSeq X sequencing platform, which is generated by human genome sequencing, has a short sequencing cycle and high throughput, and can produce at least 1.8T of data after 3 days of operation. Not only that, HiSeq X also successfully broke through the individual genome sequencing costs of thousands of yuan. With the emergence of HiSeq X sequencing platform, human whole genome sequencing has entered a new development period.
2.Experiment Process
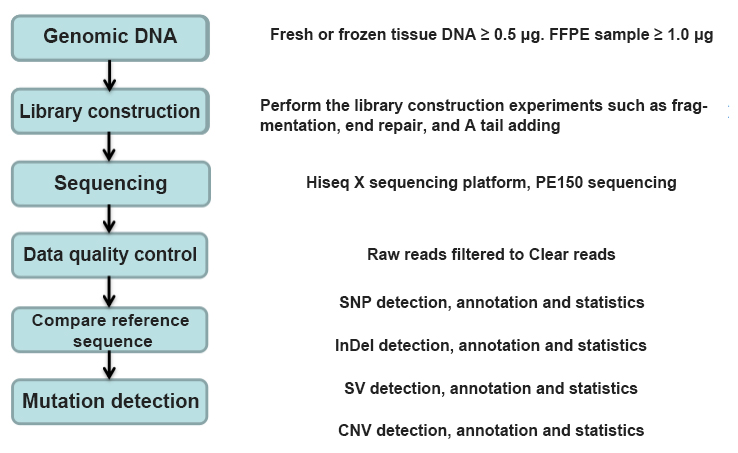
Figure 2 Whole Genome Sequencing Experimental Process
3.Technical Characteristics and Application Fields
The whole genome sequencing cycle is short, and the whole genome sequencing data can be obtained. The effective data coverage is uniform, which can be used for the detection of copy number variation (CNV) and structure variation (SV), fusion gene detection, virus integration site detection, non coding region mutation detection, etc.
Whole genome sequencing is applicable to cancer genomics, genetic disease research, drug genomics, population evolution and other research fields.
(1)In the research of cancer genome, each cancer sample presented in front of the researchers is a genome that has changed, which contains unique and unpredictable multiple point mutations, insertion, volatility, fusion, and other distortion. Since many mutations have never been observed before, and do not necessarily exist in the coding area of the genome, the whole genome sequencing is gradually considered to be the only rigorous method, and all mutations in the cancer genome can be found.
(2)In the study of genetic diseases, the whole genome sequencing can search for disease-related candidate sites or regions within the whole genome. It is especially suitable for the type of disease type with less molecular research basis or a large structural variation.
(3)In the study of human evolution, genetic information obtained by general genome sequencing can be used for screening for population evolution, population specific genes and regions.
4.Research Trend
In recent years, with the continuous development and maturity of high-throughput sequencing technology, the sequencing throughput is increasing, and the cost is greatly reduced. In addition, the importance of non coding region variation and structural variation is widely recognized, the whole genome sequencing is becoming more and more popular. From the "Human Genome Project", which cost US $2.7 billion in the 1990s, to today's "thousand yuan genome" era, whole genome sequencing has been applied to various fields, and more and more representative research articles have been published (Figure 3).
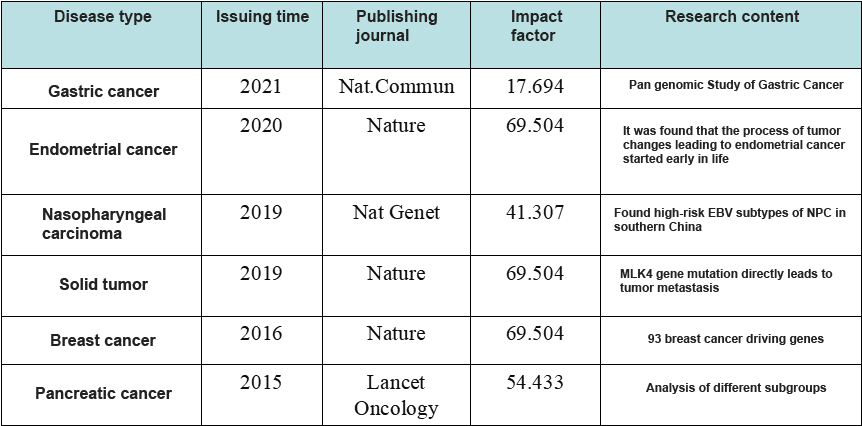
Figure 3 The Representative WGS Research Articles in Recent Years
In genome sequencing technology, the most basic and complicated operation is library preparation, which requires repeated addition of various reagents, product purification, etc., and the quality of the library plays a decisive role in sequencing results. Taking Ilumina ® Nextera XT DNA library preparation kit as an example, the overall includes a series of processes such as genomic DNA fragmentation, end repair, A-tailing addition, adaptor connection, amplification, purification, library standardization and library mixing. In this process, it need to be mixed and added various reagents, and the process of repeatedly pipetting can reach more than 600 times (take the preparation of 24 libraries as an example).
ALLSHENG Auto-NGS 100R is a fully automatic library preparation system that can meet 24 sample libraries at the same time. It only needs to be placed in the prepared nucleic acid to obtain the library product in a short time. Auto-NGS 100R perfectly replaces the tedious pipetting process, and while improving the efficiency of the construction, it also guarantees the accuracy and batch stability of the sample, minimizing the errors caused by artificial pipetting. The operation logic is clear, which greatly reduces the difficulty of use.

Reference
[1] China Birth Defects Prevention and Control Report (2012) [R]. Beijing: Ministry of Health of the People's Republic of China, 2012
[2] Liu Chunlin, Chen Peisong, He Xiaohong, et al Application of Non-invasive DNA Screening in Prenatal Diagnosis of 4723 Pregnant Women [J] China Maternal and Child Health Care, 2020, 35 (21): 4055-4059.
[3] Wu L, Wu Y, Zou S, et al. Eliciting women’s preference for prenatal testing in China: a discrete choice experiment [J]. BMC pregnancy and childbirth, 2020, 20(1): 1-8.
[4] O'Leary P, Maxwell S, Murch A, et al. Prenatal screening for D own syndrome in A ustralia: costs and benefits of current and novel screening strategies [J]. Australian and New Zealand Journal of Obstetrics and Gynaecology, 2013, 53(5): 425-433.
[5] Ashoor G, Syngelaki A, Poon L C Y, et al. Fetal fraction in maternal plasma cell‐free DNA at 11–13 weeks' gestation: relation to maternal and fetal characteristics [J]. Ultrasound in Obstetrics & Gynecology, 2013, 41(1): 26-32.
[6] Gil M M, Accurti V, Santacruz B, et al. Analysis of cell‐free DNA in maternal blood in screening for aneuploidies: updated meta‐analysis [J]. Ultrasound in Obstetrics & Gynecology, 2017, 50(3): 302-314.

 Biological Sample Preparation
Biological Sample Preparation
 Life Science Detection Products
Life Science Detection Products
 POCT Detection & Reagent
POCT Detection & Reagent
 Automation & Liquid Handling
Automation & Liquid Handling
 Laboratory Instrument
Laboratory Instrument
 Reagent & Consumable
Reagent & Consumable
 Others
Others
 OEM/ODM
OEM/ODM












 Release time:2022-11-14
Release time:2022-11-14
 Source:
Source:
 Pageviews:4862
Pageviews:4862













 + 86 571-88859758
+ 86 571-88859758 sales@allsheng.com
sales@allsheng.com



