1
What is MRD?
MRD refers to "a few cancer cells left in the body during or after treatment". At the 18th China Lung Cancer Summit Forum in 2021, an expert consensus was reached on MRD of lung cancer: MRD of lung cancer refers to that after treatment, traditional imaging (including positron emission tomography/CT, PET/CT) or laboratory methods can not be found, but molecular abnormalities of tumor origin found through liquid biopsy represent the persistence and clinical progress of lung cancer.
2
The Relationship Between ctDNA and MRD
Circulating tumor DNA (ctDNA) is cfDNA fragments originating from malignant tumors or circulating tumor cells in blood, which exists in plasma or serum. At present, ctDNA is the most common marker of MRD. Research on advanced lung cancer shows that ctDNA analysis has important clinical value in tumor gene mutation detection, disease progress monitoring, tumor load evaluation and drug efficacy prediction.
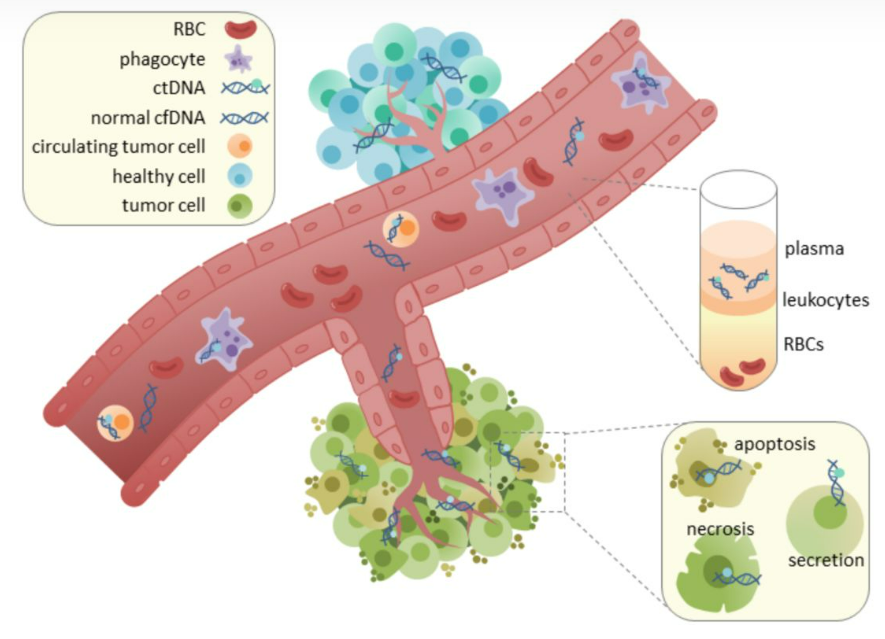
3
Detection Method
It is still an urgent clinical need to accurately detect tumors such as stage II colon cancer (CC) patients with MRD after surgery. The presence of ctDNA has a strong prognostic value for the recurrence of CC disease. In a CC clinical study, a total of 30 patients' blood samples were collected before and 4-21 days after the operation, with two 10ml tubes at each time point and large volume. At present, there are few large volume cfDNA extraction equipment on the market. Allsheng Auto-Pure 24D can extract 5/10ml of processing volume, meeting most research needs.
01 Blood Tumor Detection
One of the most effective methods to detect ctDNA is to specifically detect cfDNA from plasma to detect genomic mutations previously found in corresponding tumors. Somatic mutations (deletions in the non tumor group) were identified by sequencing tumor biopsy or surgical samples. As the gold standard for detecting MRD ctDNA, it can detect tumor variation even in a very small number of tumors.
02 PCR Detection Based on Customized Panel
Droplet digital PCR (ddPCR) is a highly specific technology that can detect a single predefined genomic variation with the VAF of 0.01%. ddPCR has the advantage of providing quantitative results, and different from quantitative PCR, it also presents a certain proportion of VAF in the same determination. Compared with other technologies, it also has high sensitivity, so a very low DNA input (~1ng) can also be used to detect a mutation.
4
Clinical Significance and Prospect of ctDNA Detection of MRD
01 One of clinical significance: prediction of recurrence of MRD after treatment of solid tumors
In some patients with solid tumors undergoing radical treatment, MRD continues to be lower than the level of imaging or physical detection threshold, which eventually leads to disease recurrence. Recent work focusing on ctDNA has shown that this analyte can be used as a biomarker of MRD in solid cancers. Although ctDNA usually represents a small part of cfDNA in plasma, it can detect tumor specific mutation, structural variation, copy number change and epigenetic characteristics through PCR or NGS, so as to identify the possibility of patients carrying MRD after radical treatment, and guide the administration of adjuvant or consolidation therapy.
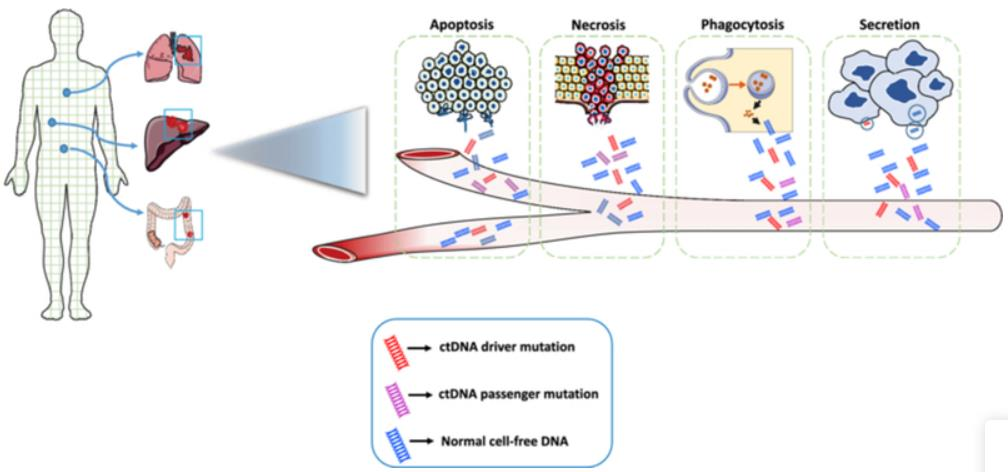
02 The second clinical significance: auxiliary treatment improves the prognosis of MRD patients
The ultimate goal of ctDNA MRD analysis is to assist/consolidate treatment to increase the possibility of cure and/or reduce toxicity. More and more published cases show that adjuvant chemotherapy can reduce the ctDNA level of MRD patients after definitive treatment. In relevant studies, 88 patients with stage III colon cancer received safe SeqS ctDNA detection after surgery and adjuvant chemotherapy. Among 18 patients with positive evidence of ctDNA MRD after surgery, 9 patients turned negative after chemotherapy, and 6 patients remained disease-free at the last follow-up. This indicates that ctDNA MRD can be used to track the response to adjuvant systemic therapy.
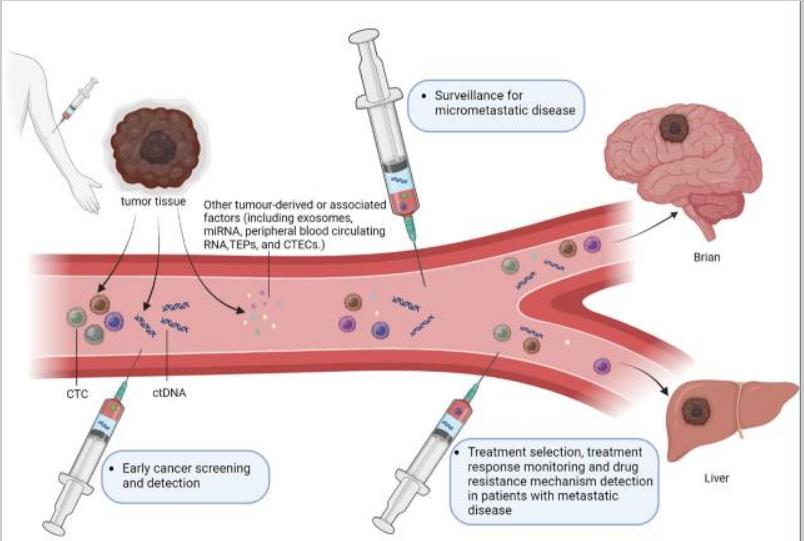
03 The third clinical significance:
It can be used for early cancer screening and detection, and can also be used to further check the abnormalities found in imaging examination.
04 Outlook:
More and more evidence shows that ctDNA can be used as a biomarker for MRD detection. The development of ultra sensitive MRD detection technology may improve the level of cancer treatment. Continuous ctDNA analysis after surgery can be used to adjust the intensity and duration of auxiliary treatment according to the results of ctDNA. In general, in the era of precision medicine, the application of ctDNA based MRD analysis has great benefits in providing clinical decision support and improving patient survival results.

Auto-Pure 24D Nucleic Acid Purification System
■The maximum sample loading volume is 4ml, and the processing system is 10mL, meeting the demand for large volume extraction
■Minimum elution volume is 50μl, which effectively ensures nucleic acid concentration
■Single tank and adapter design, 1-24 samples freely selected
Reference
1. Abbosh, C.; Frankell, A.; Garnett, A.; Harrison, T.; Weichert, M.; Licon, A.; Veeriah, S.; Daber, B.; Moreau, M.; Chesh, A.; et al. Abstract CT023: Phylogenetic tracking and minimal residual disease detection using ctDNA in early-stage NSCLC: A lung TRACERx study. Cancer Res. 2020, 80, CT023.
2. Valpione, S.; Campana, L. Detection of circulating tumor DNA (ctDNA) by digital droplet polymerase chain reaction (dd-PCR) in liquid biopsies. Methods Enzymol. 2019, 629, 1–15.
3. Hilke, F.J.; Muyas, F.; Admard, J.; Kootz, B.; Nann, D.; Welz, S.; Rieß, O.; Zips, D.; Ossowski, S.; Schroeder, C.; et al. Dynamics of cell-free tumour DNA correlate with treatment response of head and neck cancer patients receiving radiochemotherapy. Radiother. Oncol. 2020, 151, 182–189.
4. NIH. Minimal Residual Disease Definition.
5. Coakley, M.; Garcia-Murillas, I.; Turner, N.C. Molecular Residual Disease and Adjuvant Trial Design in Solid Tumors. Clin. Cancer Res. 2019, 25, 6026–6034.
6. Garcia-Murillas, I.S.G.; Weigelt, B.; Ng, C.; Hrebien, S.; Cutts, R.J.; Cheang, M.; Osin, P.; Nerurkar, A.; Kozarewa, I.; Garrido, J.A.; et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci. Transl. Med. 2015, 7, 302ra133.
7. Chera, B.S.; Kumar, S.; Shen, C.; Amdur, R.; Dagan, R.; Green, R.; Goldman, E.; Weiss, J.; Grilley-Olson, J.; Patel, S.; et al. Plasma Circulating Tumor HPV DNA for the Surveillance of Cancer Recurrence in HPV-Associated Oropharyngeal Cancer. J. Clin. Oncol. 2020, 38, 1050–1058.
8. Elazezy M, Joosse SA. Techniques of Using Circulating Tumor DNA as a Liquid Biopsy Component in Cancer Management. Comput Struct Biotechnol J (2018) 16:370–8. doi: 10.1016/j.csbj.2018.10.002.
9. Franczak C, Filhine-Tresarrieu P, Gilson P, Merlin JL, Au L, Harle A. Technical Considerations for Circulating Tumor DNA Detection in Oncology. Expert Rev Mol Diagn (2019) 19(2):121–35. doi: 10.1080/14737159.2019.1568873.

 Biological Sample Preparation
Biological Sample Preparation
 Life Science Detection Products
Life Science Detection Products
 POCT Detection & Reagent
POCT Detection & Reagent
 Automation & Liquid Handling
Automation & Liquid Handling
 Laboratory Instrument
Laboratory Instrument
 Reagent & Consumable
Reagent & Consumable
 Others
Others
 OEM/ODM
OEM/ODM












 Release time:2022-12-29
Release time:2022-12-29
 Source:
Source:
 Pageviews:4167
Pageviews:4167













 + 86 571-88859758
+ 86 571-88859758 sales@allsheng.com
sales@allsheng.com



